- Have Revised
Introduction¶
One of the areas which we’re really revolutionising with microelectronics and medical devices is the whole area of genetic technology.
You might be unaware but the whole entirety of the human genome was only first sequenced in 2001. Since then there’s been an explosion of technologies in the quest to be able to detect what is our DNA, and obviously the fringe benefits of that is you can detect DNA and RNA. This helps us with systems for detection of infectious diseases.
DNA / RNA Basics¶
DNA:¶
DNA is the molecule that carries the genetic instructions necessary for the growth, development, functioning and reproduction of all living organisms.
DNA is a double helix, the helix shape is connected either sides by base pairs
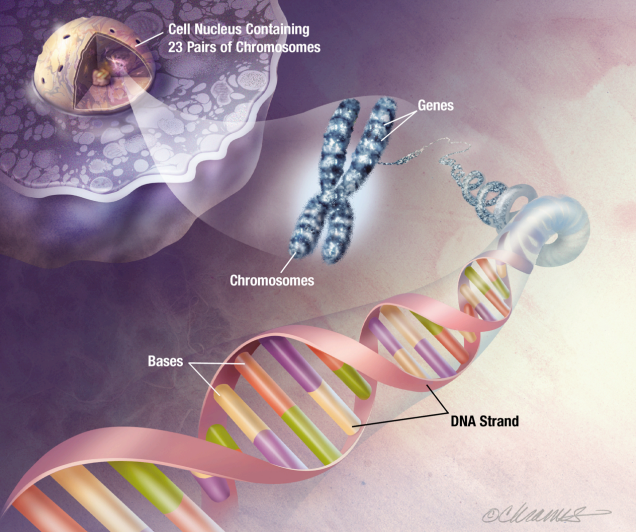
Chromosomes are long thread-like structures of tightly coiled DNA. They ensure DNA is compact and organised so it can fit inside cells and be accurately replicated during cell division. Humans have 46 chromosomes in each cell, arranged in 23 pairs of chromosomes.
Genes refer to specific segments of DNA that act as instructions to make proteins. Each gene contains a sequence of bases that determines the protein’s structure and function.
A Genome is the entirety of all genes of an organism.
DNA is stored in the nucleus of the cell, apart from in some cases the mitochondria (mtDNA)
RNA:¶
RNA is the messenger and worker for a process called protein synthesis.
Protein Synthesis occurs in 2 main stages:
Transcription
- This is the process where the information in a gene (DNA) is copied into mRNA
- It occurs in the nucleus
- This resullts in a strand of mRNA carrying genetic instructions for protein synthesis
Translation
- This is the process where the mRNA is used as a template to assemble a chain of amino acids into a protein
- the mRNA carries the genetic code from the DNA in the nucleus to the ribosome
- tRNA brings amino acids to the ribosome
- rRNA forms part of the ribosome structure and catalyzes the assembly of the protein
- This results in a newly synthesised protein which will forld into is functional shape
These resulting proteins can be used in various parts of the body for structure, transport, communication and regulation of different chemical and biological processes.
What is the difference between DNA and RNA?
| Feature | DNA | RNA |
|---|---|---|
| Primary Role | Stores genetic information | Transfers and helps decode genetic information to synthesise proteins |
| Location | Mainly in nucleus and in Mitochondria | Found in nucleus and cytoplasm |
| Strands | Double-Stranded Helix | Single-stranded |
| Bases | Adenine (A), Thymine (T), Cytosine (C), Guanine (G) | Adenine (A), Uracil (U) (instead of Thymine), Cytosine (C), Guanine (G) |
| Length | Long and stable | Shorter and less stable |
| Structure | Rigid and helical | Flexible and variable in shape |
Bases¶
As mentioned before, the DNA double helix is made up of complementary base pairs formed by hydrogen bonding. A sequence of 3 base pairs is called a codon
What are they for?
- They store genetic information - a codon in RNA corresponds to a specific amino acid in protein synthesis
- DNA replication - Each strand serves as a template and complementary base pairing ensures accurate duplication
- and many more
We have the following base pairs:
DNA Base Pairs:
- Adenine (A) pairs with Thymine (T) (2 hydrogen bonds)
- Cytosine (C) pairs with Guanine (G). (3 hydrogen bonds)
RNA Base Pairs:
- Adenine (A) pairs with Uracil (U) instead of Thymine (T) (2 hydrogen bonds)
- Cytosine (C) pairs with Guanine (G). (3 hydrogen bonds)
DNA Physical Properties¶
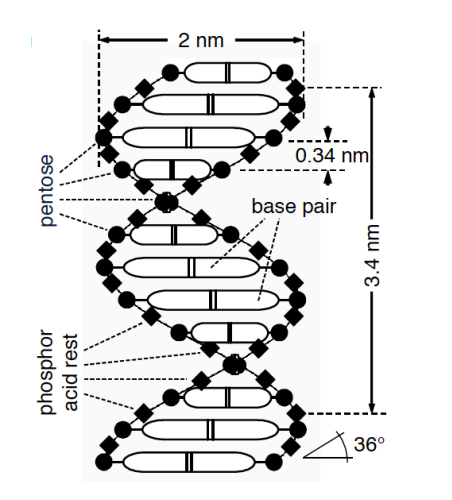
The dimensions and physical properties of DNA can be seen in this figure.
DNA physical properties:
- Negatively charged
- Mass of
- Lengths (in humans) of
DNA Sensing / Detection¶
There’s 2 types of ways to detect DNA:
DNA Sequencing
- You have a sample of the DNA and you want to figure out what the sequence of the bases are without any prior knowledge
- So you determine the order of the bases in sequence in terms of A, T, C, G in DNA molecules
DNA Hybridisation
- Here we are looking for a specific strand of DNA code, e.g ssRNA viruses like covid!
- It’s usually a highly parallel investigation of a given sample concerning the amount of specific pre-defined DNA sequences
- So not about a sequencing an unknown piece of DNA, it’s about a targeted investigation of a sample
- And DNA Hybridisation means a binding of sequence of bases
Applications¶
DNA sequencing and DNA Hybridisation can be used for a number of things:
Full Genome Sequencing - Determine the whole genome of an individual and screen for predisposition to certain disease, e.g diabetes, cancer, etc.
SNP Detection - Detecting SNPs or ‘mutations’ in our DNA, where an expected base pair is found to be another. These can determine our biological traits, or tell us if we are predisposed to certain diseases.
SNPs¶
SNPs (pronounced ‘snips’) are important because they can determine the way patients respond to drugs. To achieve an appropriate drug response a drug has to reach a minimum concentration, these drug concentrations are influenced by how the body interacts with the target protein - which are traits dictated by individual SNPs.
Detecting SNPs (Hybridisation)¶
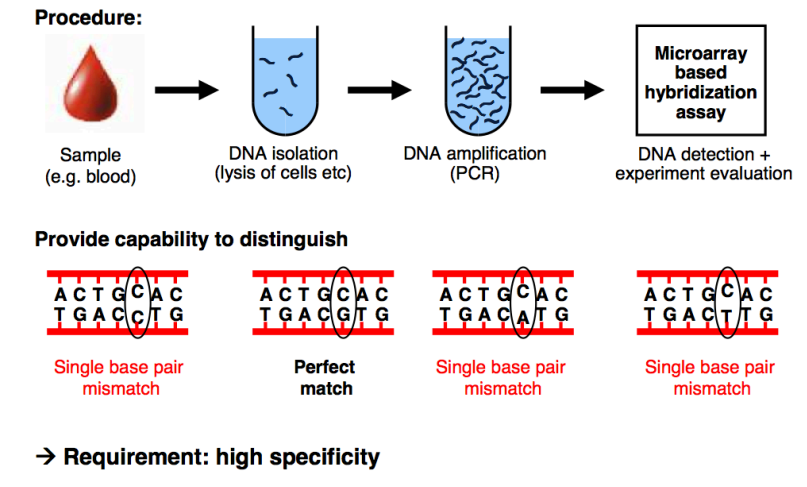
Let’s break down the procedure in this figure:
Sample Collection
- Obtain a biological sample, providing a source of DNA
- e.g blood
DNA Isolation
- Extract the DNA from cells in the sample
- This involves breaking open the cells (lysis) to release the DNA and purify it for downstream analysis
DNA Amplification
- The isolated DNA is amplified using PCR
- The PCR generates enough DNA for analysis by copying the regions of interest repeatedly
Microarray-Based Hybridization Array
- This is the detection step which analyses the DNA using a series of complementary probes to target what you’re looking for
- these probes bind to specified sequences by setting the bases, so it will bind when the complimentary pairs are found in the sequence
DNA Microarray Chips¶
So what are DNA microarray chips?
They are perform highly parallel investigation concerning the presence/absence/quantitative amount of specific DNA sequences in a given sample.
They are chips of the order mad eof glass/polymer material/Silicon. They are used for genome research, drug development and medical diagnosis.
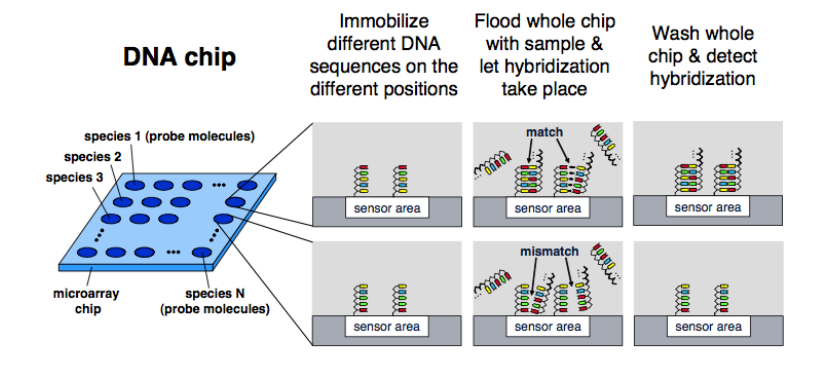
Here’s an example of an optical microarray. It’s got an array of sites and on each of those sites is where you have a sensor. You create a probe which is for the specific target DNA you’re looking for, so the complimentary pairs.
If at each of these sites, the target sequence is immobilised, the sample is flood over and if there’s a match binding occurs. Measuring when this binding occurs is traditionally done using optics, we will do it with pH and charge in CMOS.
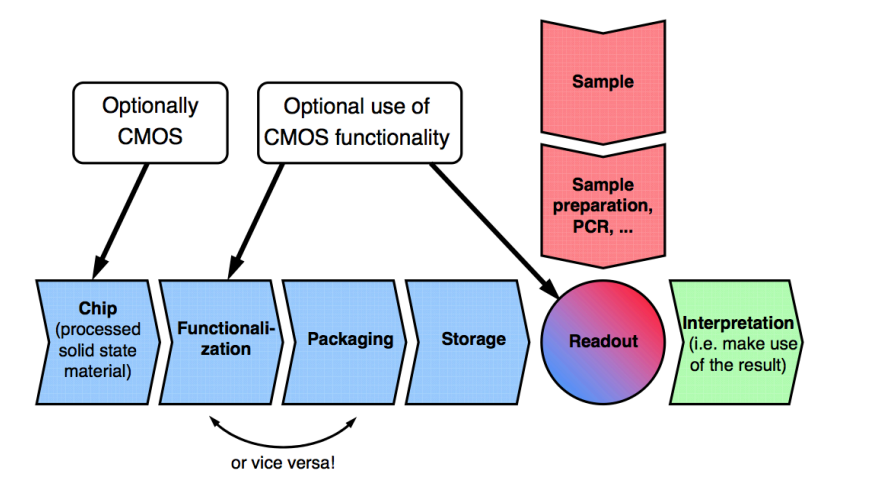
To make a microarray you need:
- A chip to mobilise the DNA, it could be CMOS or glass (depending on binding sensor methodology)
- A packaging and storing system
- A sample extraction technique to extract the DNA
- A readout to interpret the results
Here’s the application chain for DNA microarrays
Here are a couple of DNA detection methods:
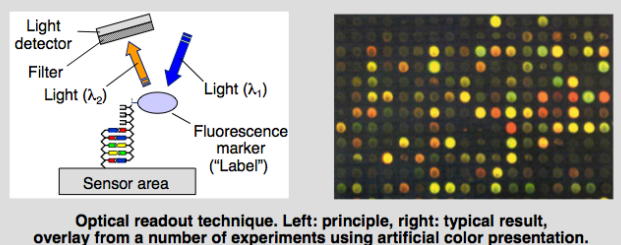
Optical based
- Using fluorescent molecules and a camera to see which DNA strands have hybridised
- Bind fluroscent marker to target, it binds if it’s complementary. You then shine light at a certain wavelength and then you’ve got a camera which can see what each site is doing.
- Light signifies the presence of the target
- Colour or intensity signifies quantity
Semiconductor based (CMOS)
- Using solid state sensors to investigate what is happening with the DNA directly above each sensor
MEMs based
- Use MEMs cantilevers which measure the weight of DNA (not very scalable)
CMOS DNA Microarray¶
We use CMOS. This is because we can have our chip, our instrumentation and we can do electrochemical detection without the need for lights or cameras.
We have a number of advantages:
- Point-of-care Suitability
- Can produce a small (handheld) device without bulky optics
- Real-Time Detection Capability
- Instrumentation is integrated directly with sensor
- This also allows for high SNR
- Implementation of large arrays
- For arrays over 100 sites CMOS integration is mandatory, because of how small we can make our sensors
- Others include:
- Decreased cost (scales with Moore’s law)
- More robust
- More user friendly
- More flexible
So how do we measure with CMOS?
We have 3 techniques, which we have seen before:
- Electorochemical
- Apply Coloumetry or cyclic voltammetry/amperometry
- Impedance based
- Measure the change in capacitance due to a binding of DNA
- Potentiometric
- Measure the pH change due to bases binding using ISFETs
CMOS Electrode Formation¶
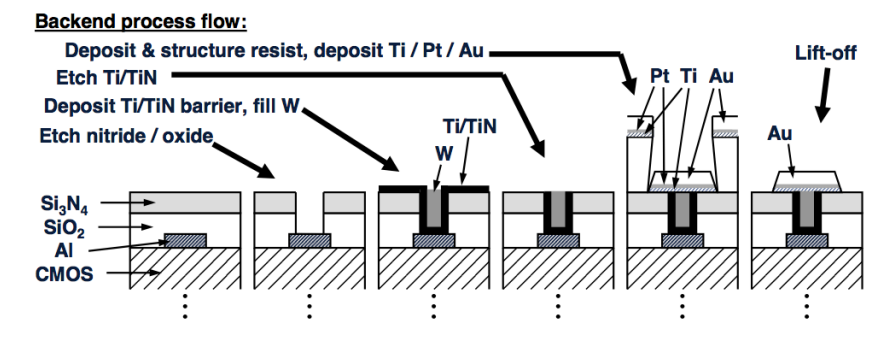
To detect in CMOS we use a silicon chip, but we need to put sensors on there to immobilise the DNA. The best sensor for electrochemical detection is an inert noble metal. We use this for electrochemical and impedance based methods, as they need formation of electrodes on the CMOS surface.
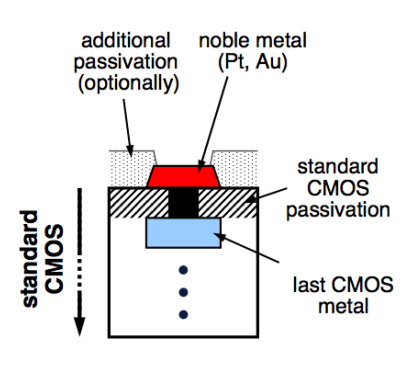
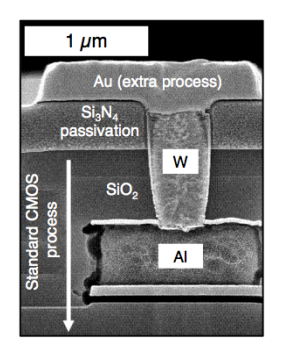
We usually use the gold or platinum, and to put it down on the chip we use what’s called a lithographic process. So you take your CMOS chip and you use an acid to burn through the top, protective dielectric layer to create a contact with the metal below. This process is called etching. You then deposit a titanium barrier on that so that the noble metal and the rest bind nicely. Once in equilibrium, you can put your gold or platinum down.
Here’s an example of this process of Au’on’Standard’0.5μm,‘6“‘CMOS’Process’using’ standard’lithography’.
Electrochemical Detection¶
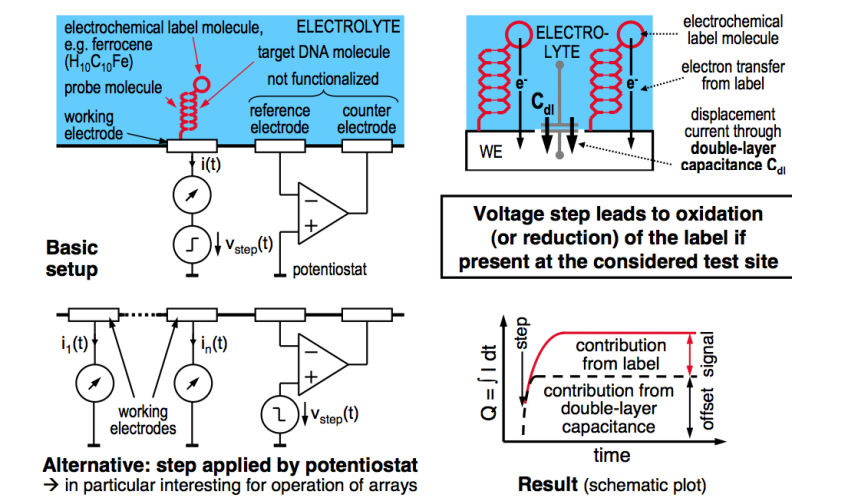
As previously discussed we can get a redox potential and current flow. So in this instance, we need something to cause a redox reaction when the DNA binds.
For these purposes we use Ferrocene (), it’s a good redox molecule.
We label the target DNA with the Ferrocene, if it binds to the DNA probe, we hit the redox potential and then the potentiostat measurement of the redox current will tell us it’s bound.
We can have an array of these working electrodes and a common potentiostat to put the redox potential.
Also because we only want to determine if it’s there, not quantify how much it is, so we can just use a step increase in the potential
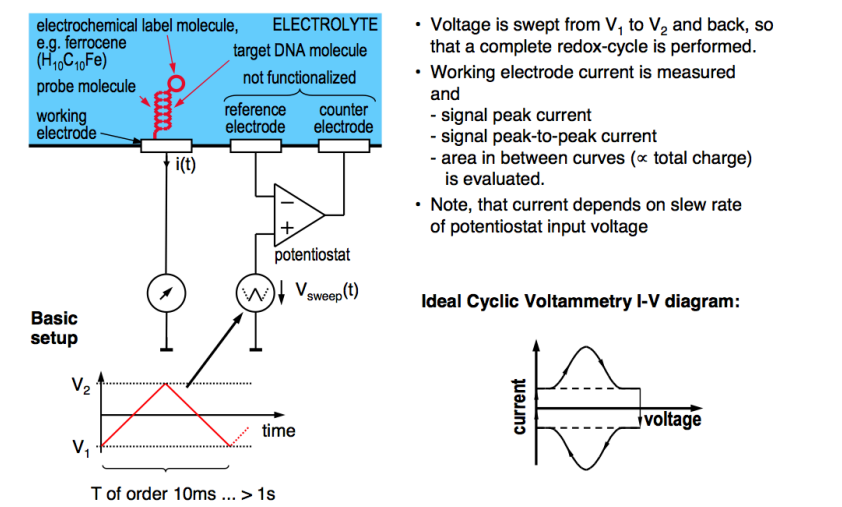
Cyclic Voltammetry¶
We can also do CV, as discussed in the past. We do a voltage sweep up and down and at the redox potential we get a peak current, which signifies the presence of the binding.
Four Electrode System¶
We can also do some more inventive stuff. Here’s an example of that, we have 2 working electrodes, one has the immobilised DNA probe, the other doesn’t. This allows us to measure it differentially.
- We use the same potentiostats and voltage generation
- Then we can measure the currents in each
- Then we can find the difference, this essentially allows us to reduce common mode noise and improve SNR
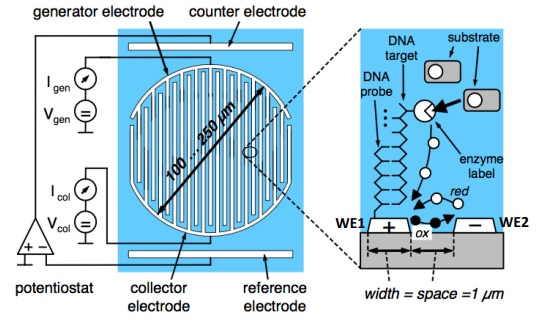
Here:
- WE1 - Target DNA molecule labeled with enzyme molecule
- WE2 - Contains no DNA and can be used as a reference for differential measurement
And here you can see the output. We can see that the slope in the derivative () signifies the presence of the target DNA. We also can see the elimination of electrochemical crosstalk.
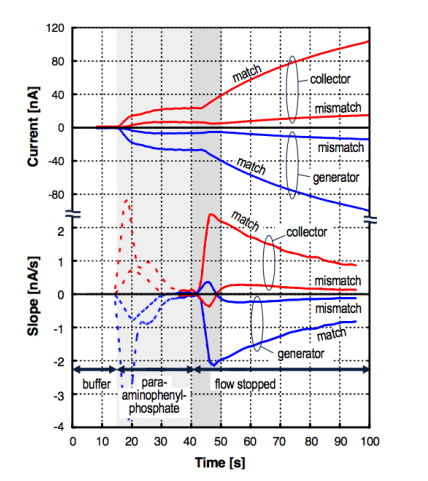
Note: The dashed lines represent the start of the reaction so we ignore that
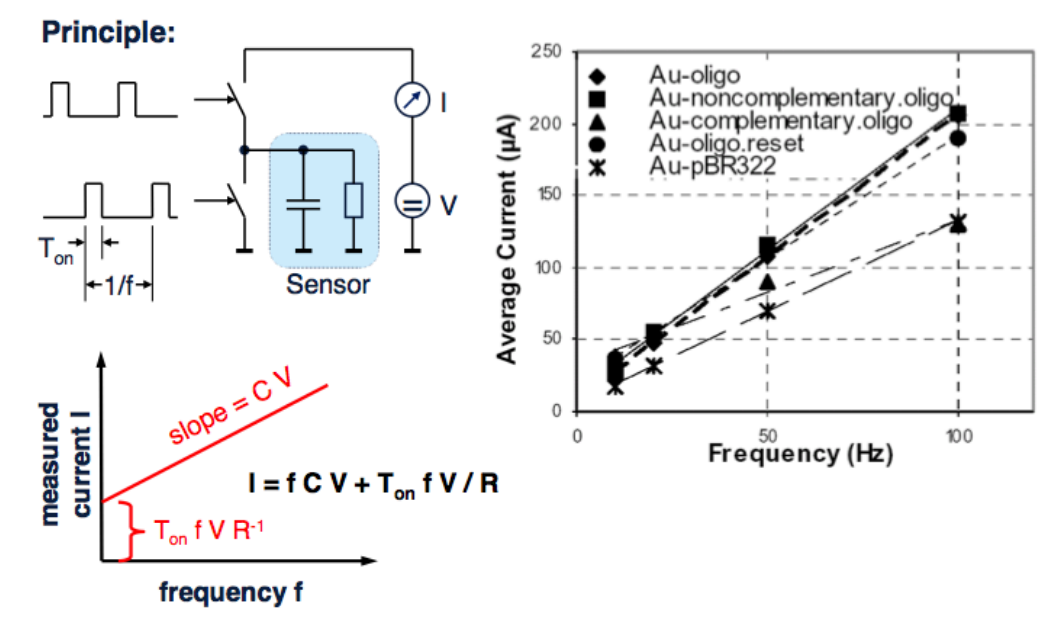
Impedance Based Detection of DNA¶
Recall that DNA has a negative charge, this means inherently when binding happens you’ll have more charge than when binding doesn’t happen. This means we can effectively measure that as an RC circuit.
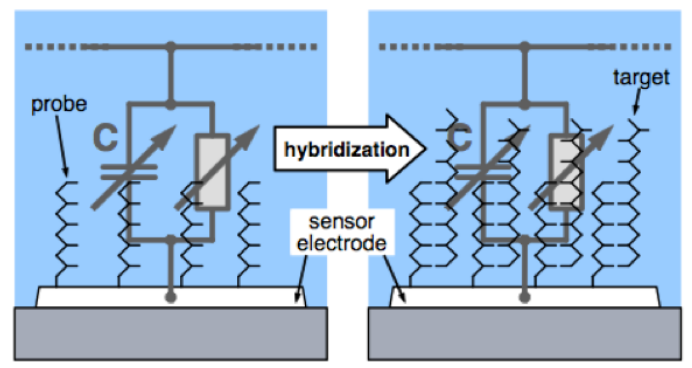
Here’s an example where you have a sensor, an RC and 2 switches pumping current in and out at a fixed frequency. So you’re charging and discharging that network with a fixed frequency. The output is a linear slope according to the provided equation.
From this equation we can extract the capacitance, which tells you exactly how much DNA there is.
pH Based DNA Detection¶
To make this process scalable and easy, we don’t want to bind the DNA to the sensor, it would be useful if it could just float on top. And we don’t want to use any light or sophisticated molecules, requiring us to pre-treat the sample.
We want a label free approach that could just take the DNA, and as it binds to its complementary it detects it.
We note the following:
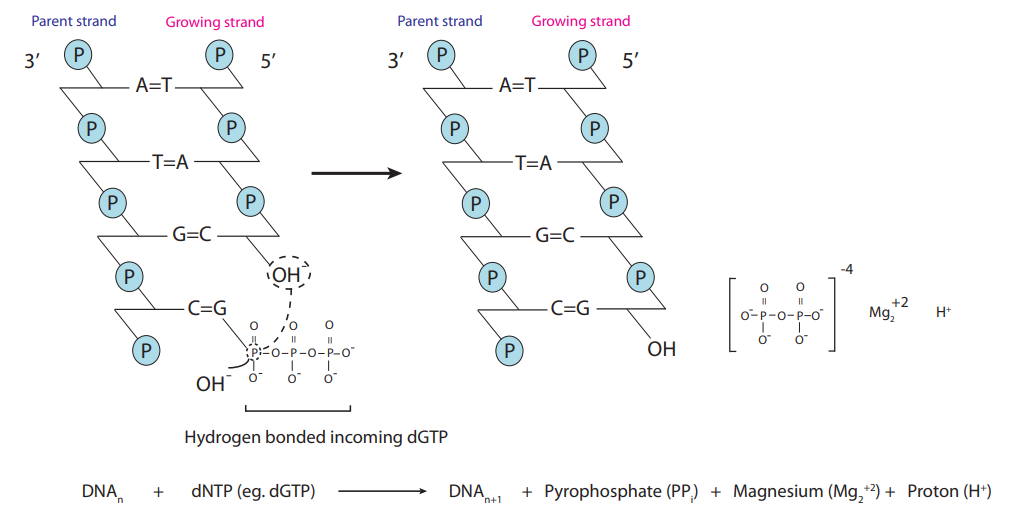
- When DNA binds in the presence of an enzyme called pyrophosphate, it does an extension and releases a pyrophosphate molecule and a proton.
- This tells us that when DNA binds, it releases a Hydrogen ion (proton) and therefore the pH will decrease
- Therefore if we have a pH sensor we can detect this process, we can do this with potentiometric methods - using ISFETs
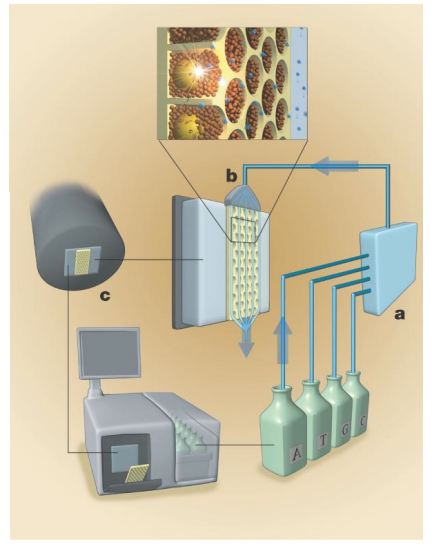
DNA Sequencing¶
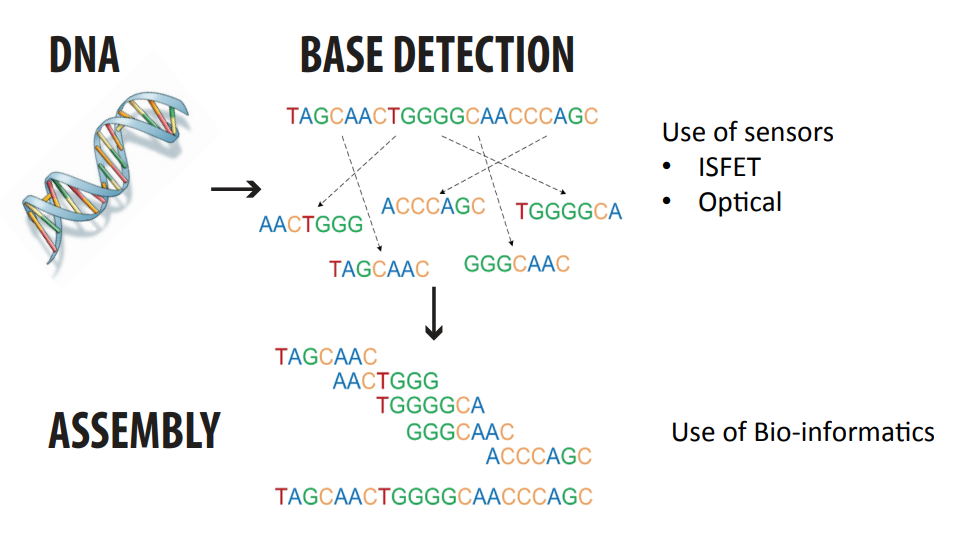
We want to take the human genome under investigation, this means evaluating 3.2 billion base pairs.
We do this by chopping them up randomly into more manageable chunks of 100-1000 bases - these are called fragments. These have overlaps.
We then immobilise them on magnetic beads, putting each bead in a well on top of a sensor.
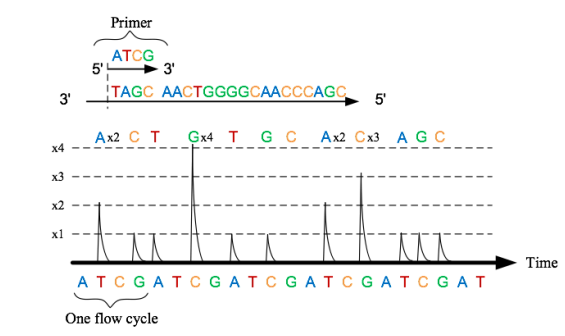
We detect the bases of the fragments by a method called pyrosequencing. A base pair match causes a change in pH which we can detect with ISFETs.
We then have an assembly algorithm that puzzles the fragments back together, it also detects overlaps in primer DNA.
Below we can see the magnetic beads in the well, note that the honeycomb structure is the most compact, allowing us to be most efficient in space.
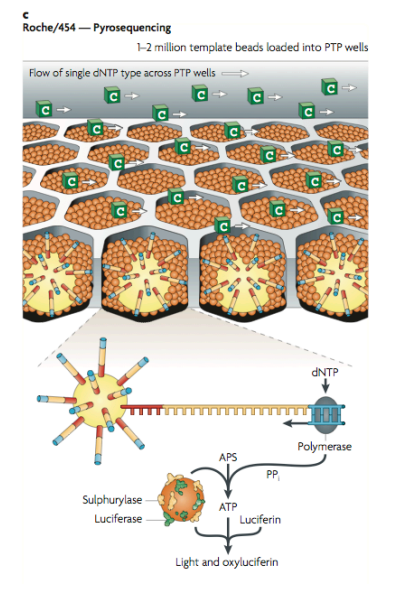
Below we can also see the use of pyrosequencing. WE have 4 bottles, in each bottle we’ve got nucleotides A, T, C and G. We sequentially insert the bases one by one, if it binds we get a signal, if not we wash and pump in the next. This means we can determine what nucleotide is in the well. This signal that we observe is generated from our ISFET array, detecting the change in pH.